The "Design new Primers" tool in Geneious provides all the necessary options required to create primer/probe sets suitable for Taqman or quantitative PCR (qPCR) assays.
If you are designing standard qPCR primers then don't include the probe primer.
First the user needs to consult the literature and select suitable design rules for their qPCR or Taqman assay. Design rules may differ depending on the polymerase/buffer system used for the assay. For example, appropriate design rules might be:
1. Primer Tm Criteria:
Forward and Reverse Primer Tm should be around 58-60 °C. Tm of the primers should be equal.
2. Probe Tm Criteria: (Taqman only)
TaqMan® probe Tm should be 10 °C higher than the Primer Tm.
3. Primer & probe %G+C Content:
Primer %G+C content should be 30-80%.
4. Primer & probe GC Clamp:
The total number of G's and C's in the last five nucleotides at the 3' end of the primer should not exceed two.
5. Primer length Criteria:
All primers should be 15-30 bases in length.
6. Primer runs and Repeats (Max Poly-X):
The primers/probes should not have runs of identical nucleotides greater than 4.
7. Primer G/C distribution:
There should be more Cs than Gs, and not a G at the 5' end.
8. Amplicon Length:
The amplicon size should not exceed 400 bp and ideally should be 50-150 bases.
Geneious can design qPCR and Taqman primers for all rules with the exception of rule 7, but this is easy to check visually via the Annotations table (see below).
To create qPCR and Taqman primers:
Select your target sequence, on the Toolbar go Primers -> Design new primers:
Use something like the following settings.
Set Task to Design New:
Check the options to design Forward and Reverse primers.
Check the option to design a DNA probe (Taqman only)
Set Task to Generic:
Set Product size Between to 50 and 150
Set Optimal Product Size to 125
Set "Number of Pairs to generate" to 5-10. This will give you a selection of primer sets that you can check to confirm at least one set conforms with Rule 7.
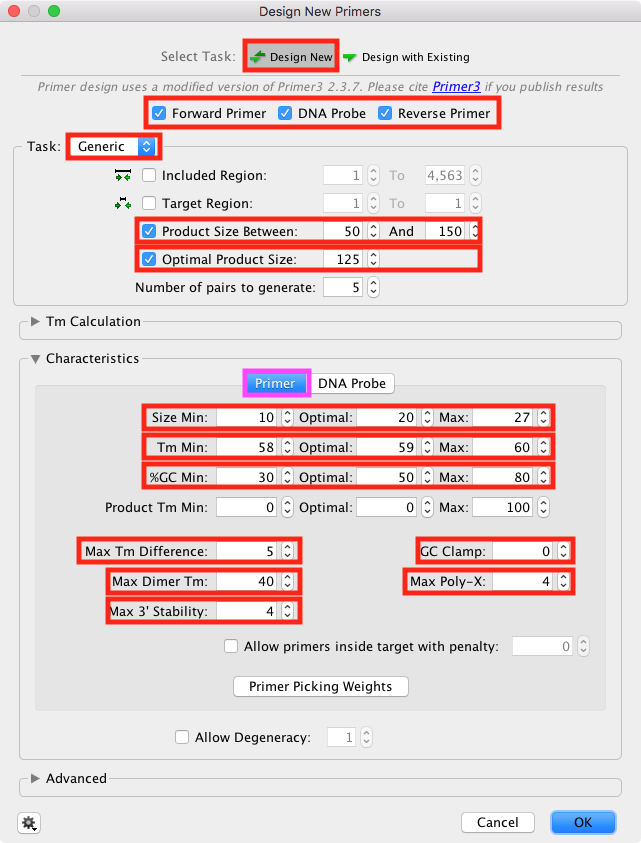
Set the Primer settings in the Primer tab
In the Characteristics -> Primer pane (highlighted magenta):
Set a broad size range
Set a tight Tm range 58°C to 60°C, optimum 59°C
Set a %GC range 30°C to 80°C, optimum 50°C.
Set Max Tm difference to 5°C
Set "GC Clamp" to 0 (this will prevent primers ending in G or C)
Set "Max Poly-X" to 4 (this will exclude primers with longer homopolymer regions)
Set "Max 3' stability" to 4 (this will favour higher A/T content at the primer 3' end)
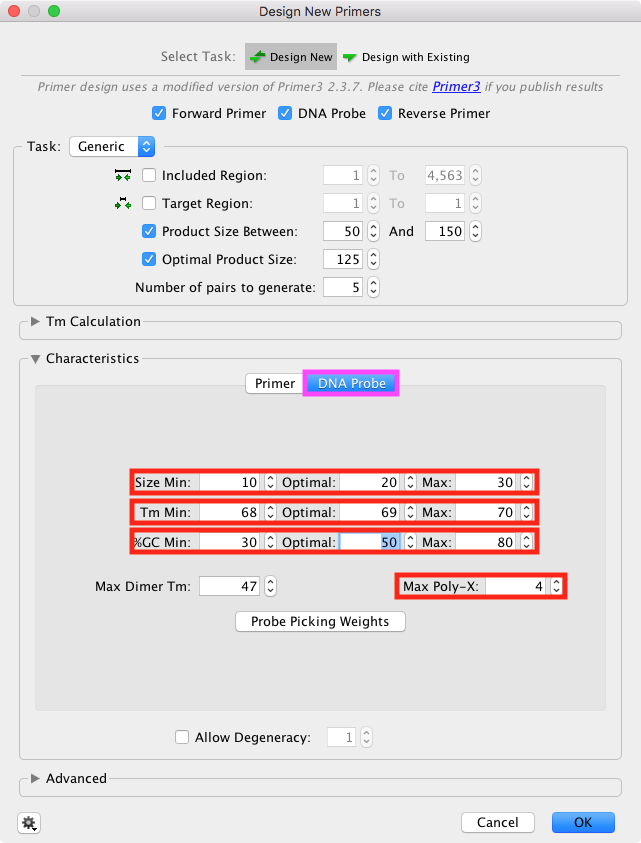
Set the Probe settings in the Probe tab (Taqman only)
Then click on the DNA Probe tab (marked magenta) and set the Tm settings for the probe.
Set the Tm range 68°C-70°C, optimum 69°C (10°C higher the the primer Tm's)
Ser Max Poly-X to 4 (this will exclude primers with longer homopolymer regions)
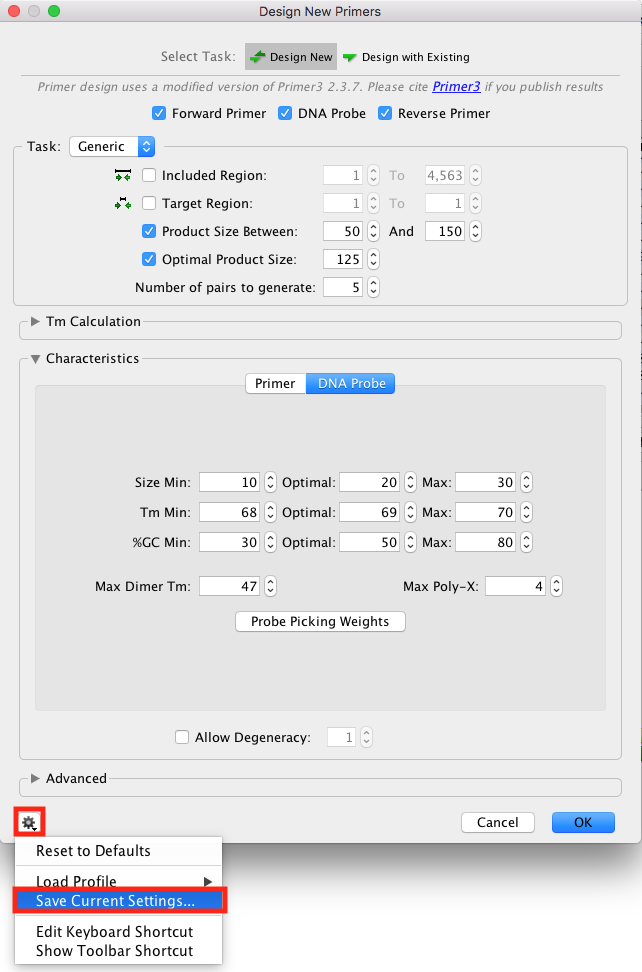
Save your design Settings
Before you run the "Design New primers" tool, you can save your qPCR or Taqman-specific design settings for future use:
Click on the cog in the Bottom left corner of the settings window, go -> "Save Current Settings".
To access the settings in future, go Primers -> Design new Primers, click on the cog icon, and choose Load Profile.
Press OK and the tool will design the number of primer/probe pairs specified. The new primers will be annotated onto the target sequence.
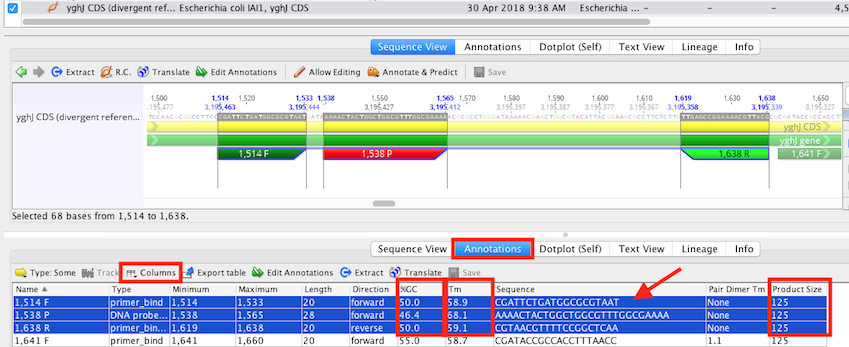
Check and validate the Primers sets via the Annotations Table
You should then check the primers in the Annotations Table to confirm they conform to the design rules.
Click on the Annotations tab in the sequence viewer:
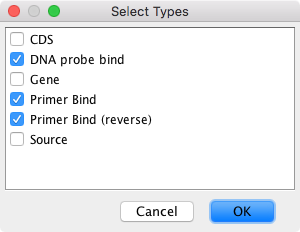
Set Type: to show Some, set to display Types "DNA probe bind" (Taqman only), "Primer Bind" and "Primer Bind (Reverse)"
Use the Columns button/Drop down menu to display relevant meta data, for example, check options to display Sequence, "Product size", Tm columns.
Confirm there are no extended G+C clamps at 3' end of the primers and that the primers/probe conform to rule 7, where there should be more Cs than Gs, and not a G at the 5' end(see below, marked by red arrow).
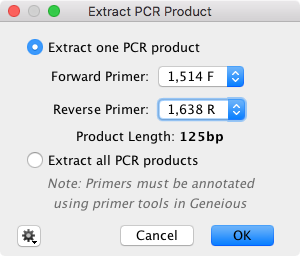
Finally, on the Toolbar, go Primers -> "Extract PCR product" to extract the chosen product using the chosen primer set.
Select the extracted product, view the statistics (%) Tab associated with the extracted PCR product and check the extracted PCR product has an acceptable %G+C.
Once you have a set of primers/probe and a final product that conforms to your qPCR or Taqman design criteria, select the primer annotations on the query sequence, then hit Extract to extract the primers/probe to new individual primers sequences.
You can then order the primers from your favourite DNA synthesis service provider, making sure you specify the appropriate fluorophore/quencher additions to the 5' and 3' ends of the probe.