In Geneious versions 6.0 and above, you can automatically annotate plasmids with common promoters, terminators, cloning sites, restriction sites, reporter genes, affinity tags, selectable marker genes, replication origins and open reading frames. To annotate your plasmids, select Annotate and Predict → Annotate From Database.... In Geneious Prime 2020.0 and earlier, this will annotate plasmids with features found in the database used by PlasMapper.
Geneious Prime 2020.1 replaces the Sample Documents/Plasmapper features folder with a new locked database of Reference Features called Geneious Plasmid Features. This database contains an expanded and revised list of common plasmid features and is now the default database for the Annotate from database tool. The contents of the Geneious Plasmid Features database can be viewed in the Sources panel under Reference Features.
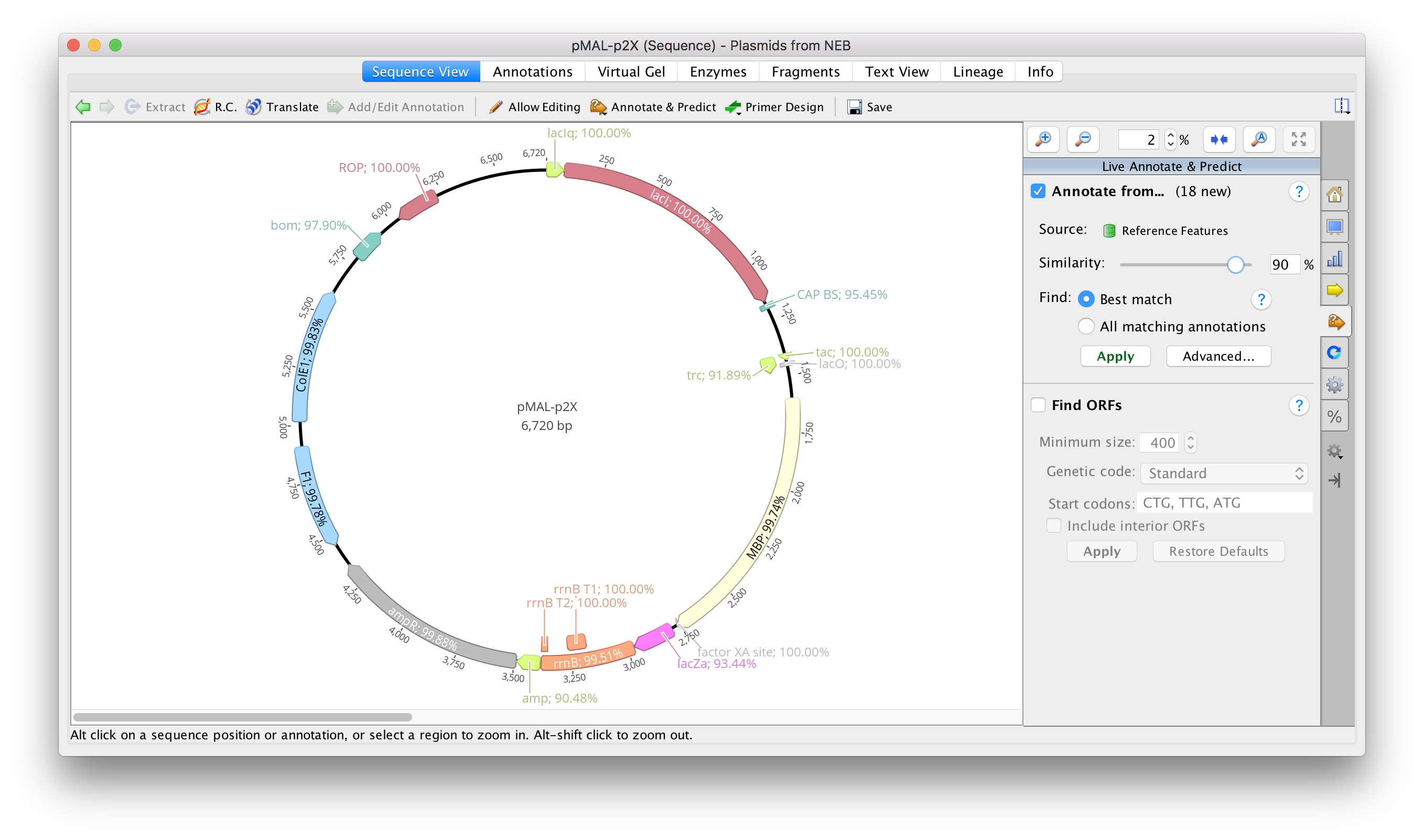
To annotate your plasmids, ensure that Reference Features or Geneious Plasmid Features is set as the Source in the Annotate From Database set up.
Alter the Similarity to set the minimum percentage identity for transferring the annotations. Insertions and deletions count as mismatches, while ambiguous matches are counted as a partial mismatch (e.g. N vs. A is 0.75 of a mismatch). Similarity is calculated along the full length of the annotation, so if only half of your sequence covers the annotation the similarity will be 50%.
To transfer the annotations to your sequence, select Apply, and then Save to save these changes to your document. The annotations will not be permanently applied to your sequence until you do this, and will appear faded - as shown in the screenshot above.
The Advanced... button also provides a selection of advanced settings that you can choose to alter how your sequences are annotated.
For more information on using the Annotate from Database tool, including how to use your own custom sequences as an annotation database, see Annotating your sequence from a custom annotation database.