This file cannot be submitted. Tbl2asn produced a submission file with no sequences.
The technical details may show the following:
Tbl2asn Output
[NULL_Caption] This copy of tbl2asn is more than a year old. Please download the current version.
[tbl2asn 25.6] SeqID lcl|xxxx is present on multiple Bioseqs in record
The first part of the error (this copy of tbl2asn is more than a year old) can be ignored, this does not prevent submission and you do not need to download a new version of tbl2asn.
The error is listed on the last line and indicates an invalid field has been chosen as the Sequence ID - "xxxx" will be replaced by the invalid field in your error message. If two or more of the sequences you are submitting have the same Sequence ID you will get this error. The Sequence ID must contain a different value for each sequence in your submission, so that each sequence can be identified during the submission process before a unique accession number is assigned. You will need to check that you have chosen an appropriate document field for the Sequence ID in the Genbank Submission setup window.
Inconsistent taxnames
This error can occur when the organism name specified in the Submit to Genbank window does not match the organism label in the source annotation on your sequence. This often occurs if you have inadvertently transferred a "Source" annotation from another sequence to your sequence. This source annotation may not be visible if the check box next to this annotation type is not ticked in the Annotations panel. To fix this error, you will either need to delete the source annotation or correct the organism label on the source annotation so that it matches your organism name exactly.
Illegal start codon used
NCBI will automatically generate the amino acid translation for any CDS annotation using the transl_table and codon_start labels on the annotation. This error message occurs when this translation does not start with the start codon, M, and you have not specified that the CDS is partial. To correct this, you will need to change the transl_table and/or codon_start label using the Edit Annotations window so that the first codon is a start codon. Alternatively, if this CDS is a partial CDS sequence, open the Edit Annotations window and double click on the annotation interval. In the window that pops up, specify that the 5' end of the annotation is truncated.
Missing stop codon
This error message occurs when the automatic translation of a CDS annotation does not end with a stop codon and the CDS is not labelled as partial. Check that the frame and translation table for this annotation are correct, and, if not, change these properties. Alternatively, if this CDS is a partial CDS sequence, open the Edit Annotations window and double click on the annotation interval. In the window that pops up, specify that the 3' end of the annotation is truncated.
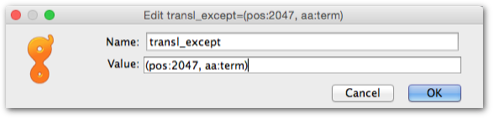
Some mitochondrial genomes contain CDS's that do not include a DNA-encoded stop codon. A stop codon is created after transcription by polyadenylation, where the terminal 'T' or 'TA' of the transcript is combined with the poly-A tail to form a 'TAA' stop codon. These non-standard CDS's require an extra property attached to the CDS to be acceptable for submission. You need to add the property transl_except to the CDS. The figure below shows the format for this property, pos:XXXX indicates the position of the terminal 'T' nucleotide of the CDS, aa:term indicates that two A's should be appended to the 3' terminus. If the CDS ends TA, then the Value: would be (pos:1..2, aa:term) where 1 is the sequence position of the 'T' nucleotide and 2 is the position of the 'A'.
Missing qualifiers or features
Annotation qualifiers should be written in lowercase letters, otherwise they will not be recognised. For example, if you write the "product" qualifier, which is required for CDS and other amino acid feature (e.g. tRNA) annotations, with a capital "P" (Product) you will get error messages such as:
Missing encoded amino acid qualifier in tRNA feature, or
No protein BioSeq given FEATURE:CDS and
Expected CDS Product absent
"Sequence x is lacking a reference" How do I create a reference?
This error message occurs if you are submitting an alignment to GenBank and one (or more) of the sequences in your alignment is not linked to an original sequence document. This will occur if you have imported the alignment into Geneious, but may also occur if you have edited a sequence in the alignment and chosen not to apply the changes to the original sequence when saving (this will break the link to the original sequence).
Sequences in the alignment that are linked to an original sequence document will have a blue arrow to the left of the sequence name in the alignment. Clicking this link will take you to the linked original sequence document. For sequences in the alignment that are missing a linked original sequence document, it is possible to create a reference sequence without realigning the sequences. To do this, select the sequence name in the alignment (this will select the entire sequence) and click the Extract button. This will create a new sequence document. Any additional information you need to add to the submission (for example collection data) should be added to this new document.