The mappers in Geneious Prime will only work with DNA sequences.
You can map peptides to a reference protein sequence using multiple alignment with non-standard settings as follows:
1. Select all of your peptides and your reference protein sequence.
2. Go Align/Assemble - Multiple Alignment.
3. Choose the Geneious Alignment tool, click More/Fewer Options and set Gap extension penalty to 0 and Refinement Iterations to at least 3.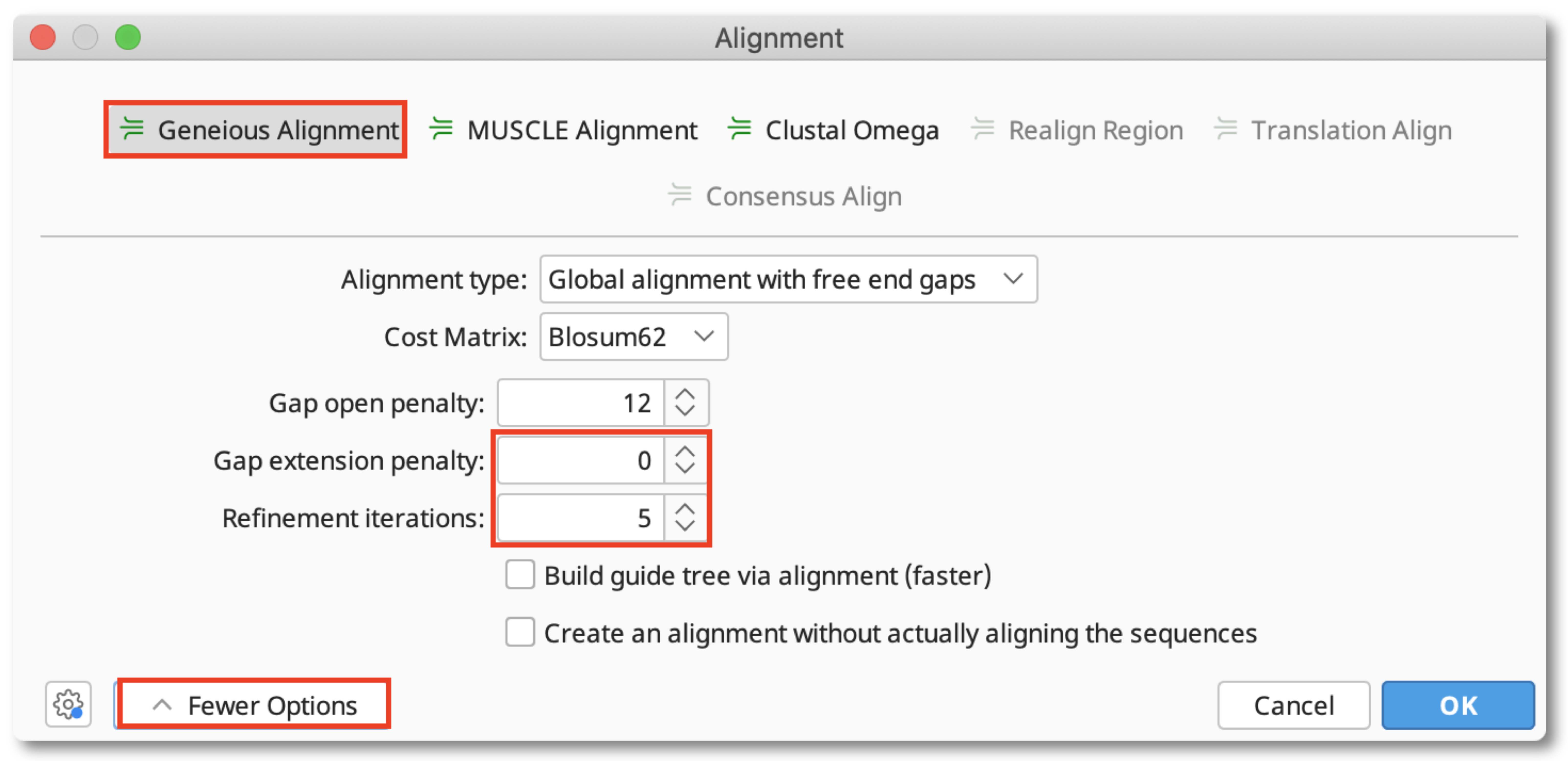
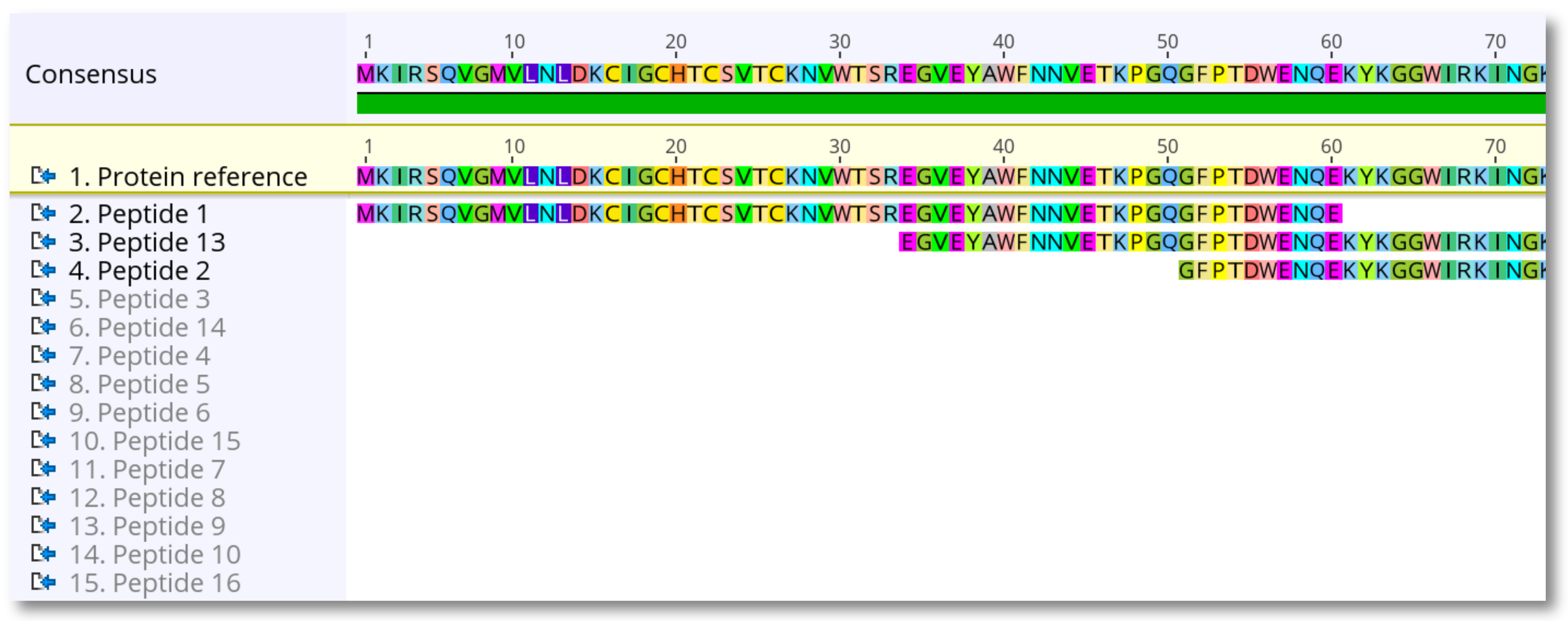
Click OK and the peptides should align correctly to the reference protein.
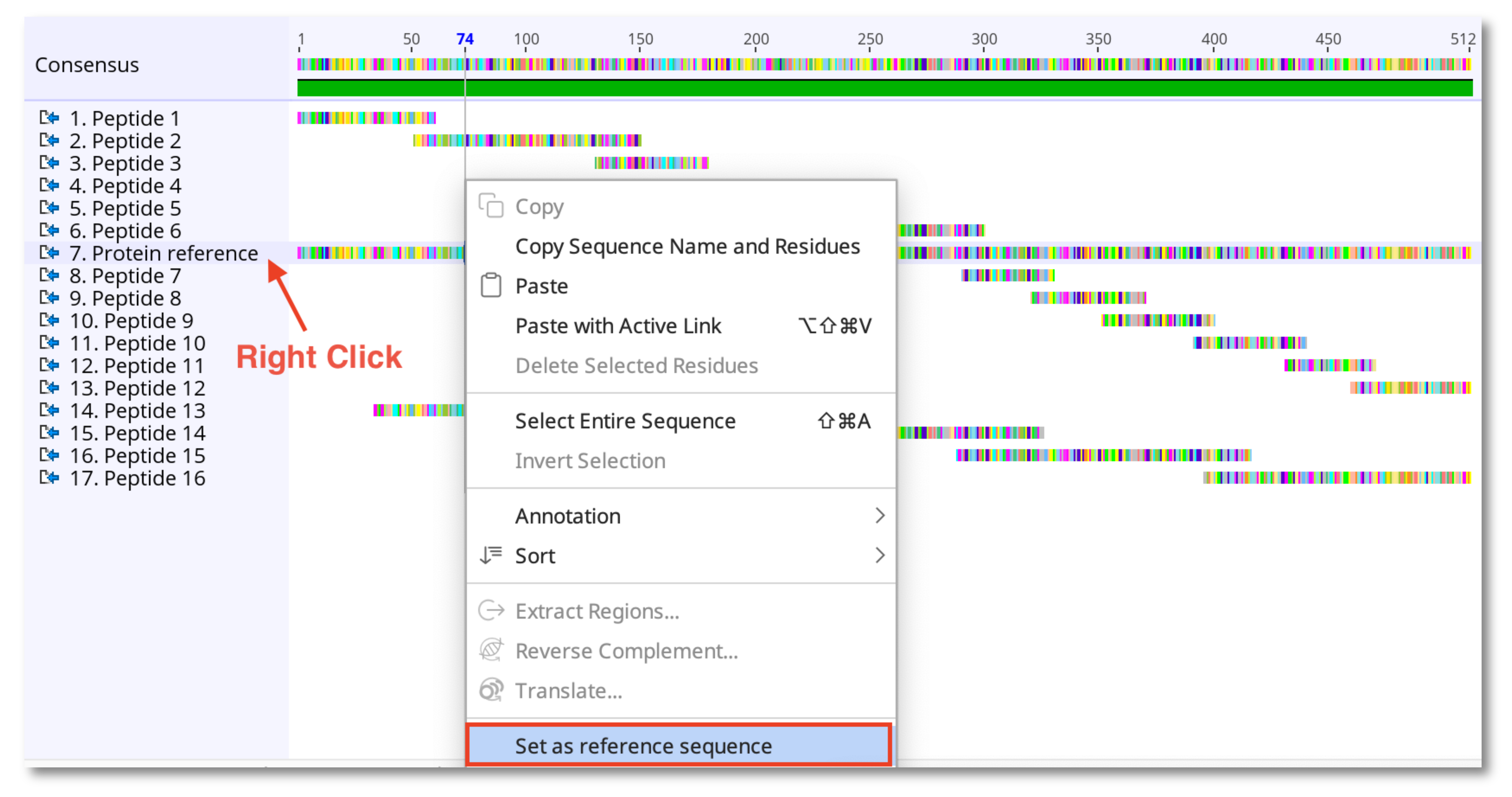
4. Select the new alignment, right-click on the reference sequence and choose Set as reference sequence. The reference will be highlighted yellow.
5. If required, click and drag the Name of the Reference sequence to move it to the top of the alignment.
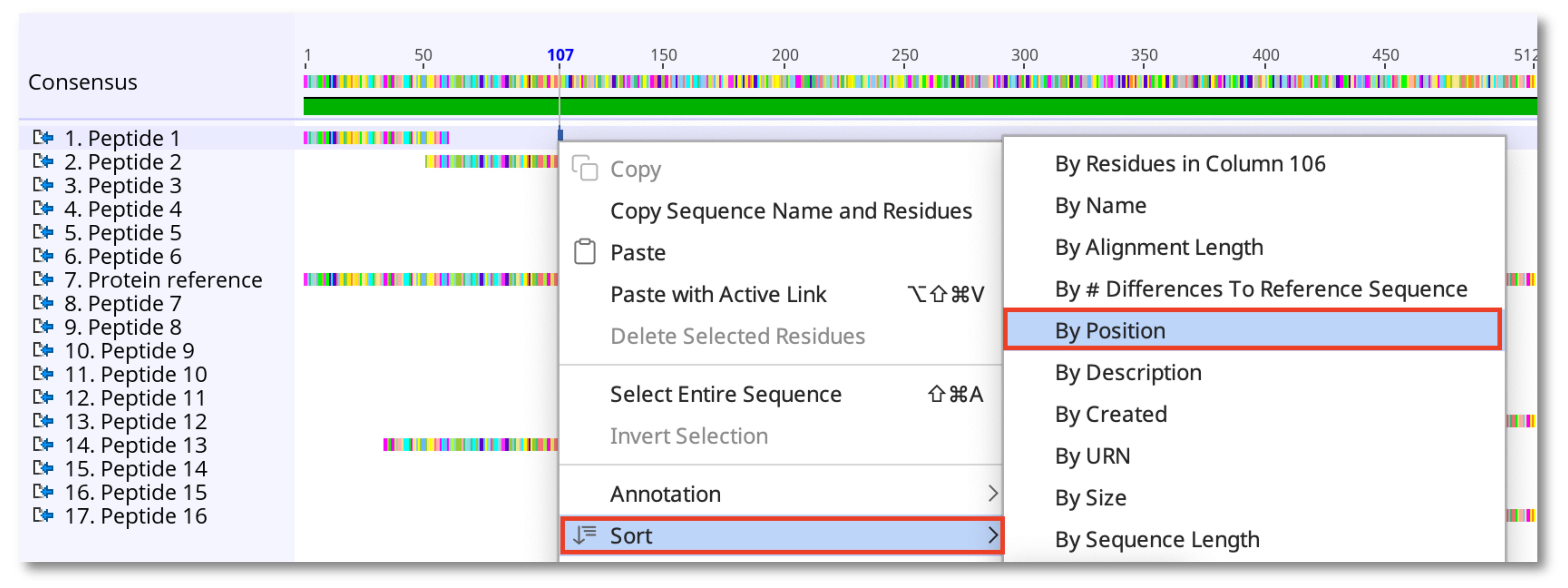
6. Right click on any sequence in the alignment and choose Sort - By Position to order all peptides sequentially based on their start coordinate.
Save. You should now have the equivalent of a map to reference assembly of peptides to a protein sequence. By default, the consensus sequence will be based on the peptide sequences only.