The Geneious de novo assembler is intended for use with bacterial genomes and small eukaryote genomes. For larger eukaryote genomes we recommend using Velvet, which is available as a Geneious plugin. For more information on which assembler to use see the following Knowledge base article Which-de-novo-assembly-algorithm-is-best-for-my-data-
Storage: The Geneious de novo assembler has very high disk space requirements, so for large assemblies 10’s to 100’s of gigabytes temporary storage may be required. Storing your database on a solid state drive (SSD) will greatly improve speed and performance.
Processor: The Geneious de novo assembler is partially multithreaded so adding processors will speed it up somewhat. As with most things computing, the faster the processor and more cores the quicker things will be.
RAM: How much RAM you require will depend primarily on the size of your data set and the quality of your dataset. The graph below provides a rough guide for the minimum and maximum amount of RAM you might need depending on your data set size and type.
The Geneious de novo Assembler always provides a minimum/maximum estimate of how much RAM will be required for assembly of a selected dataset and will not run if it expects it will not have enough RAM to complete the assembly. If in doubt, simply select your data and start the Assembler to see predicted memory requirements
General rules of thumb for RAM requirements:
- Assembling data with higher coverage depth will require more RAM – aim for coverage between 50 and 100x
- Assembling lower quality data, with more miscalls, indels and gaps, will require more RAM
- Doubling the size of your dataset (total nucleotides) will roughly double RAM requirements
- For illumina data roughly 1 GB of RAM will be required to assemble a data set of 1 million reads (with an average read length of 100 nucleotides)
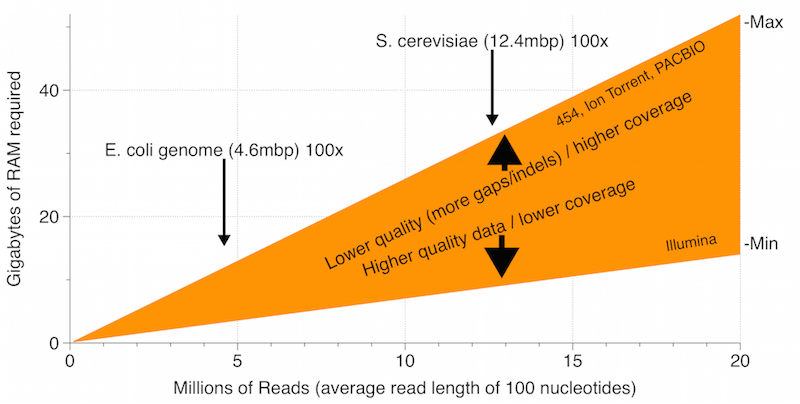
Note that the Geneious de novo assembler will allow you to trade off RAM requirements for speed (Choose Custom Sensitivity & More Options to reduce RAM requirements). The lowest setting will reduce your RAM requirements by roughly half.
How long will my de novo assembly take?:
The time required for the Geneious de novo assembler to complete will depend on your hardware, the size of your dataset, and the settings used for assembly. Reducing the “Sensitivity” setting will increase the stringency of the assembler and reduce the time required to complete assembly.