In Geneious R10 and later, sites containing gaps can be removed from an alignment by using the Mask alignment function under the Tools menu. This tool provides the option to either annotate these regions within your existing document, or to create a copy of your file that has these sites removed.
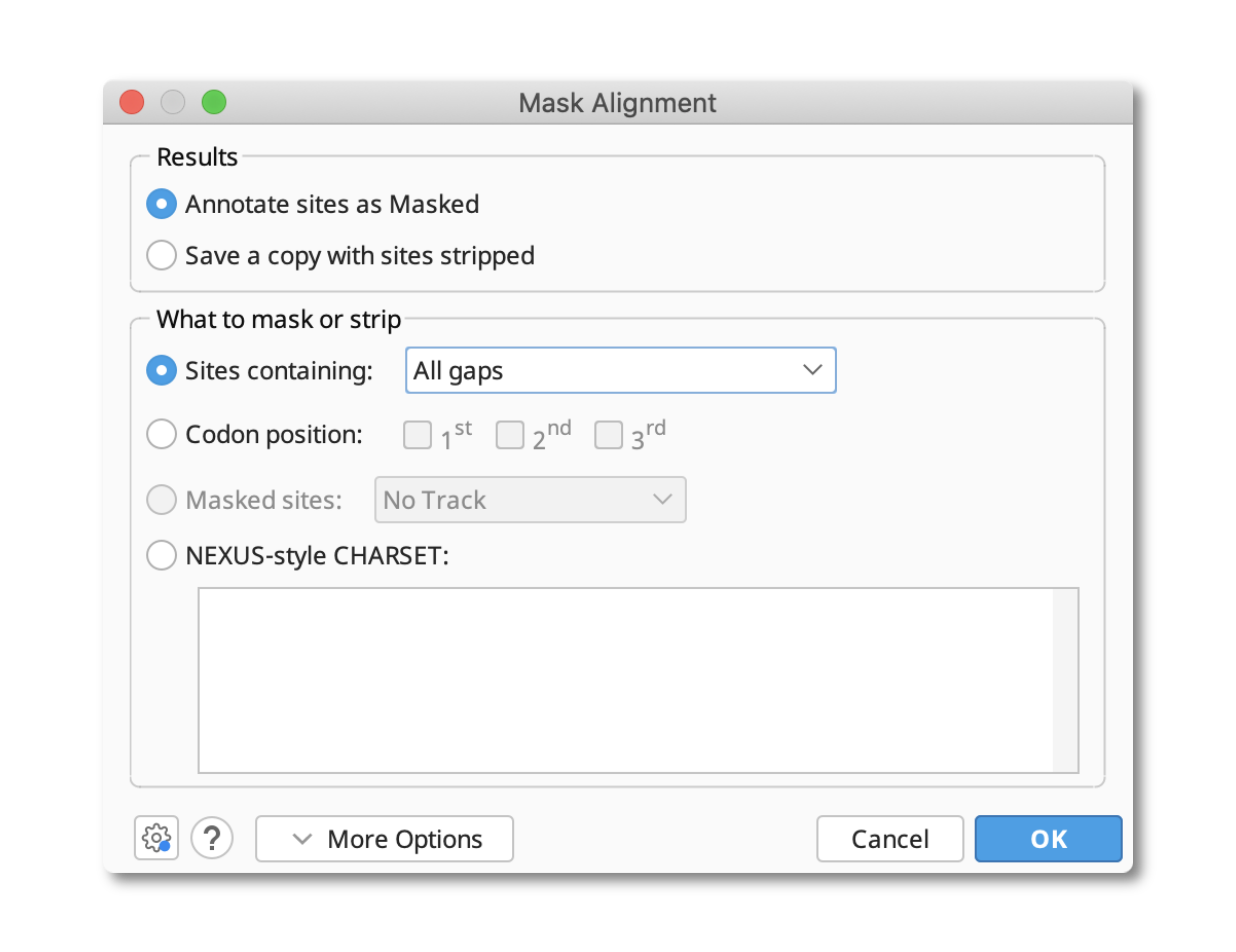
Under the Sites containing option it is possible to specify the types of gaps that you want removed from your sequence using the following options:
- All gaps: will masks or strip all sites containing only gaps. Sites containing gaps as well as residues will be preserved.
- Gaps (%): Masks or strips all sites containing at least the specified percentage of gaps (inclusive). Sites containing less than this percentage of gaps will be preserved.
- Any gaps: Masks or strips all sites containing at least one gap.
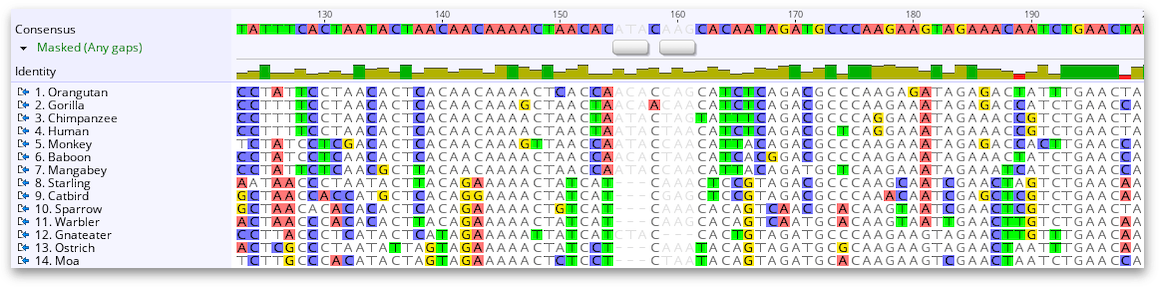
If you choose to Annotate sites as Masked, you will see a separate annotation appear on your document. This annotation tells you that the sites are masked and the type of masking that has been performed (any gaps in the below example).
These masked sites can then be omitted from phylogenetic analyses by specifying Exclude masked sites in the tree building menu.
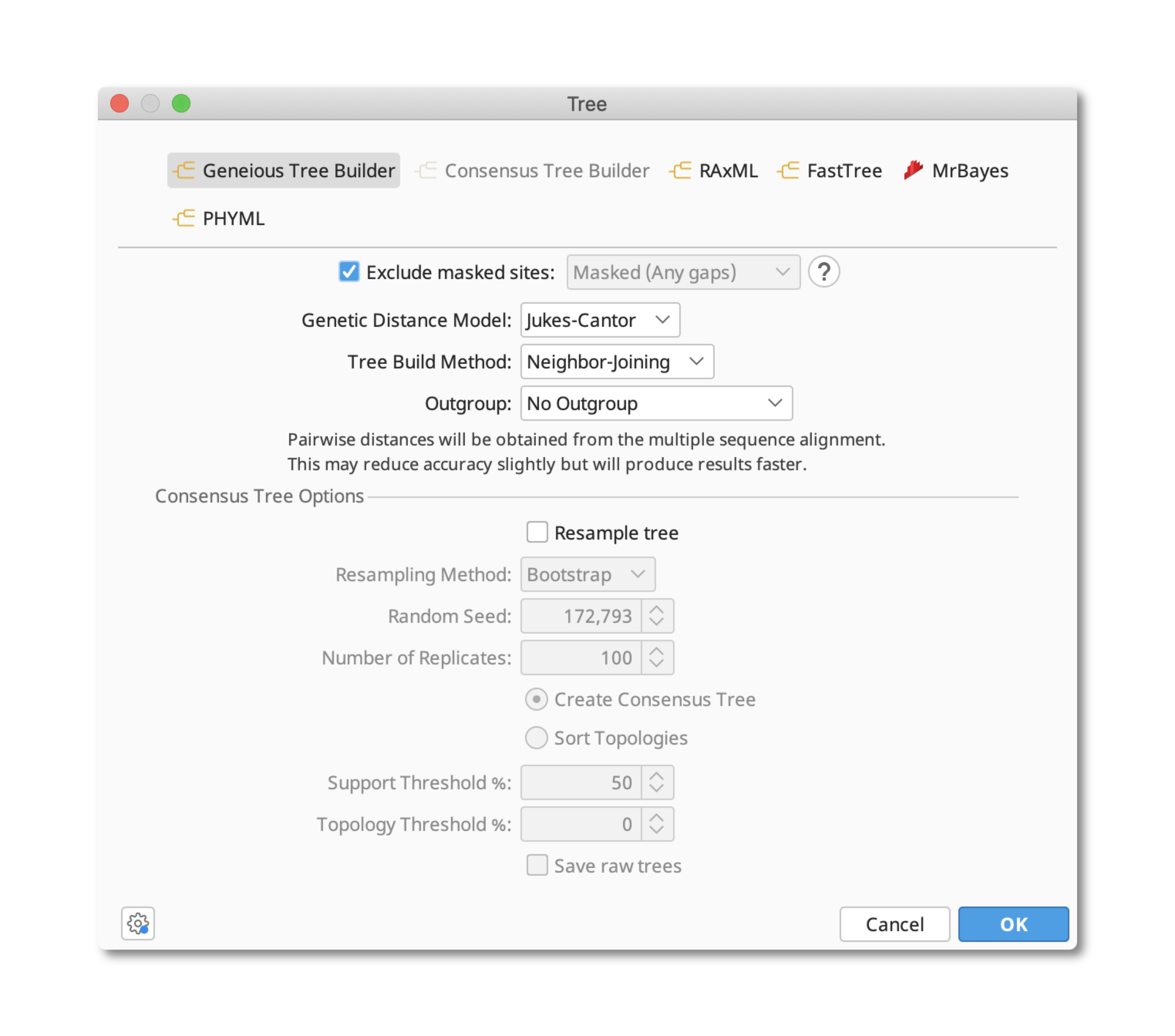
If you have chosen to save as a copy with the gaps stripped, then you can directly use the newly saved copy document to create your tree without specifying the exclusion of masked sites.
In Geneious R9 and earlier this function is found under Tools -> Strip Alignment Columns. This function does not allow you to annotate your existing document with masked sites, and instead will automatically create a copy of your alignment with the gaps stripped based on the settings you have specified.