In Geneious Prime 2019.1 onwards the Text View tab is able to display your sequences or alignments in customisable plain text which can then be exported or copied, or in a rich (formatted) text format which can be copied and pasted into a text editor or Microsoft Word document.
Further improvements have been made to this tool in Geneious Prime 2019.2. If you are using Geneious Prime 2019.1 then please update to version 2019.2.
To use "Custom text view " feature, select a sequence or an alignment and switch to the Text View pane.
Single sequences
With a one or more sequences and/or sequence lists of the same type (DNA or protein) selected, switch to the Text View pane and use the Format: dropdown menu to select whether to view in GenBank, Custom or original plain view (Don't reformat).
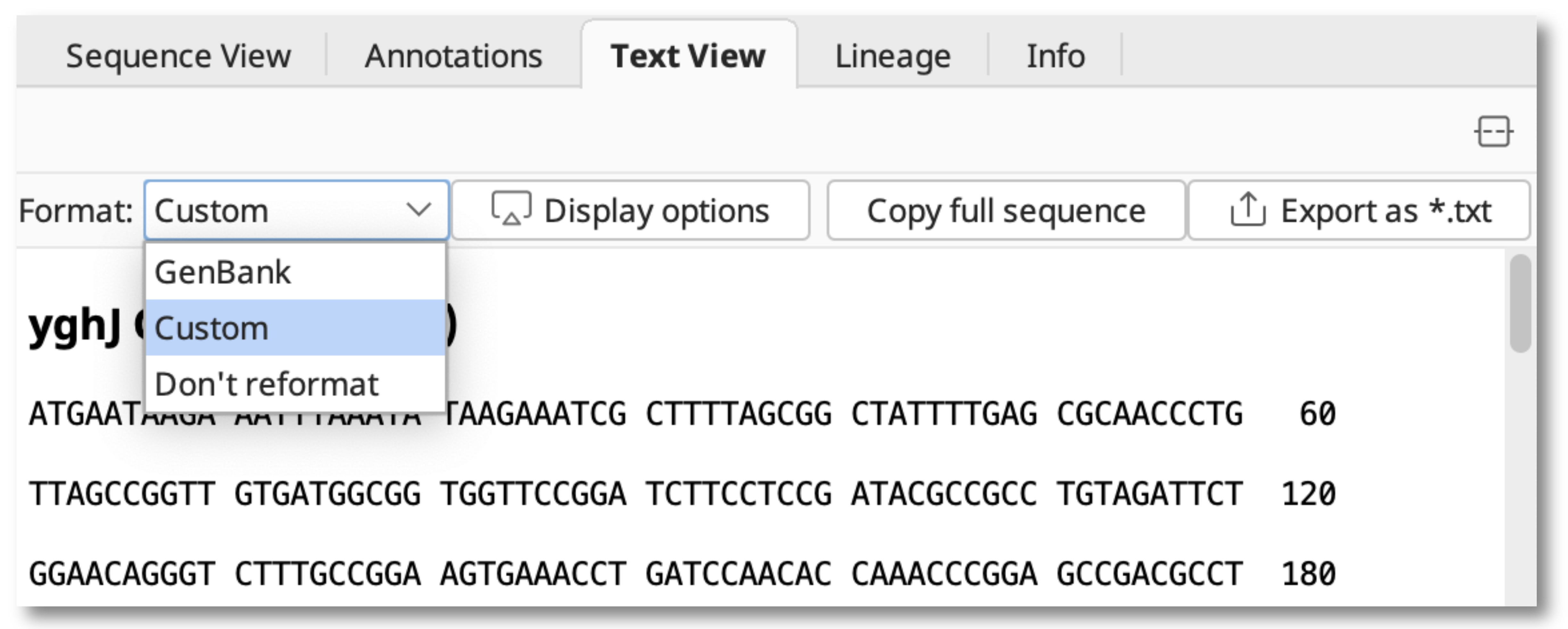
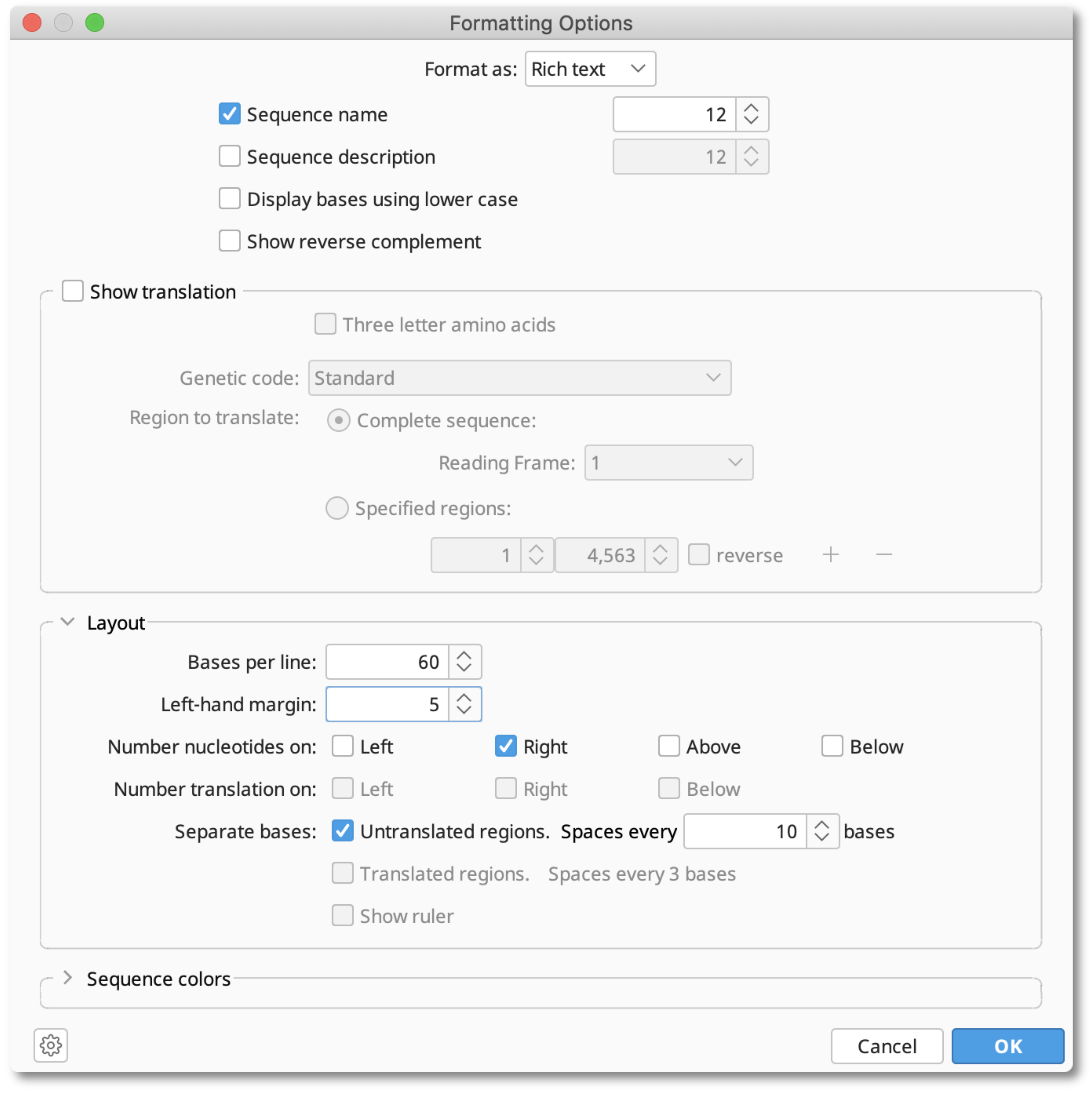
Select Custom and hit the Display options button to show the Formatting Options dialog window and choose to display in plain text or rich text format.
You can also choose to display the sequence name and description and whether the bases are in Upper (default) or lower case. To display the complementary reverse strand check Show reverse complement.
You can choose to Show translation for the complete sequence on a specific frame, or the translation across multiple ranges. Note that each CDS must be in the forward orientation. If the selected sequence has CDS annotations, and you select these Annotations in the Sequence view or Annotations table, then these regions will be automatically available to display as "Specified regions"†.
Under Layout, you can set Bases per line, set numbering/numbering position, margin width and ruler settings. You can break the sequence into blocks by checking and defining the option Separate bases: (ruler cannot be displayed if Separate bases is on).
If you choose rich text format, then you can also use the Sequence colors option to highlight one or more regions in the sequence or alignment with color.
Alignments
Select a single alignment file, switch to the Text View pane, and use the Format: dropdown menu to switch between the Custom view or the original plain view (Don't reformat).
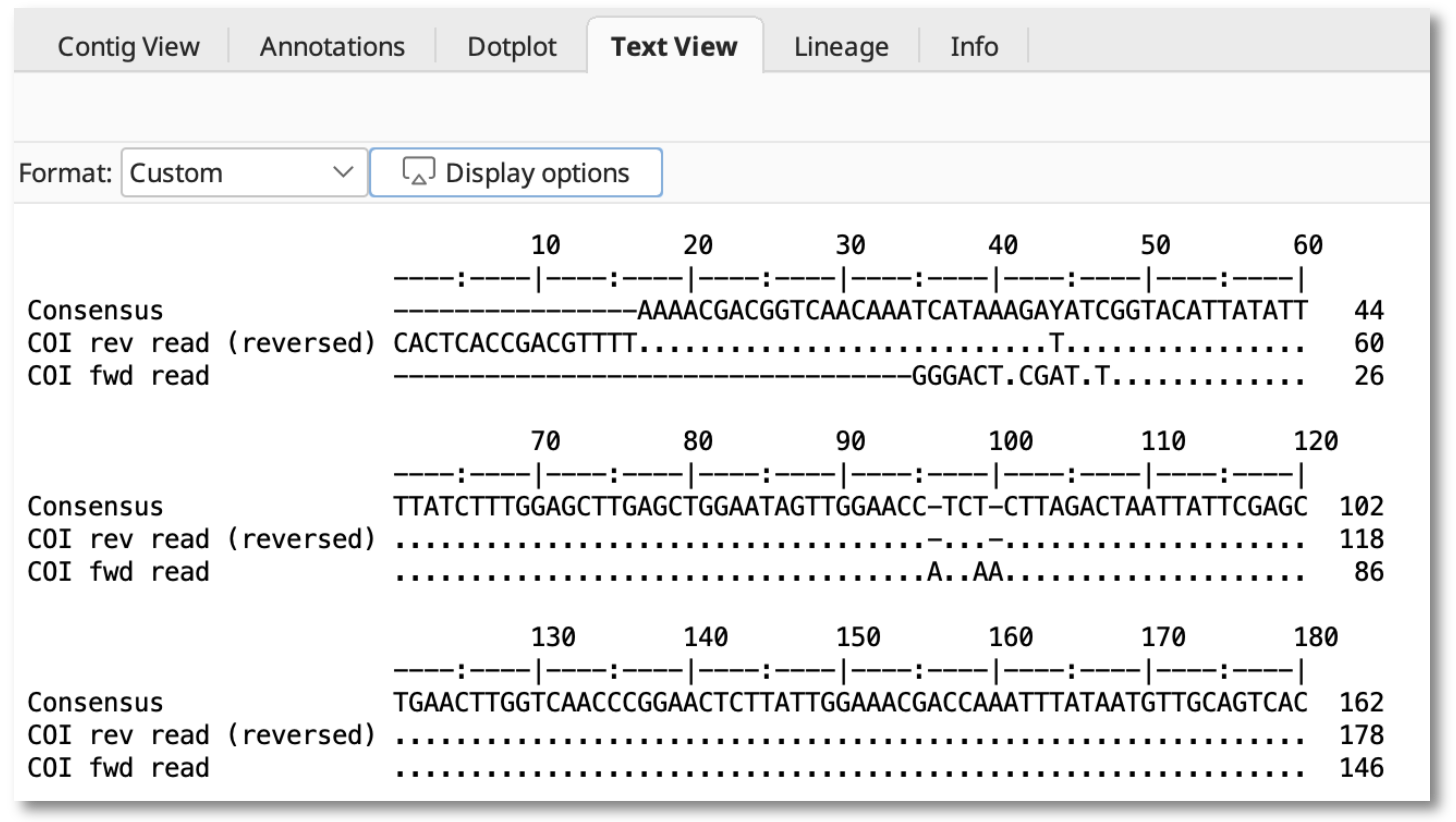
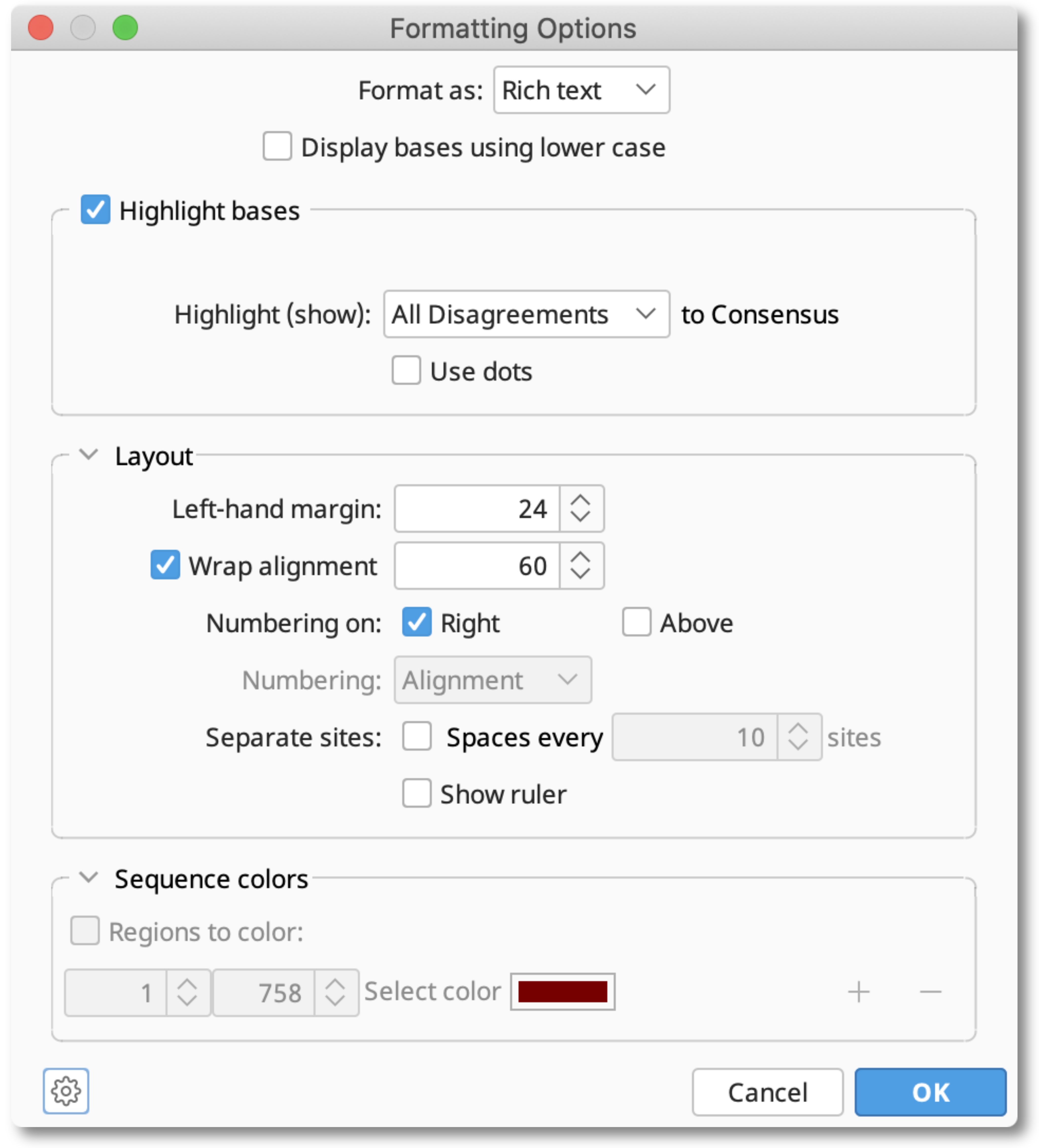
Select Custom and use the Display options button to show the Formatting Options dialog window and choose to display in plain text or rich text format.
The Highlight bases section allows you to show disagreements, agreements, similarities and dissimilarities to the Consensus.
You can also set the Layout and Sequence Colors in the same way as for a single sequence.
Exporting your text
Use the Copy full sequence button to copy the displayed text (with color formatting) to the clipboard, or use the Save as *.txt button to export the displayed text (without formatting) to a text file.
---
† If your CDS has more than one exon (a multi-interval annotation) and your exons start or end after the first or second codon positions then you will need to manually adjust the start and end coordinates for each exon to set the break to fall after the third codon position to ensure correct translation is displayed.